Accessing and Utilizing Pretrained LLMs: A Guide to Mistral AI and Other Open-Source Models"
This section explains how to access various pretrained LLMs like Mistral AI and BioMistral using the HuggingFace Hub. It highlights configurations for efficient GPU use, model-specific chat templates, and the cleaning process for evaluation.
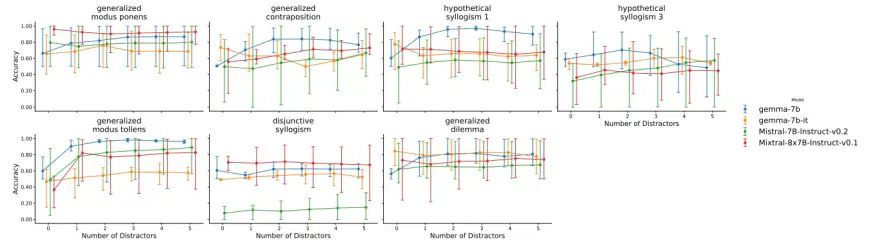
Table of Links
\
A. Formalization of the SylloBio-NLI Resource Generation Process
B. Formalization of Tasks 1 and 2
C. Dictionary of gene and pathway membership
D. Domain-specific pipeline for creating NL instances and E Accessing LLMs
H. Prompting LLMs - Zero-shot prompts
I. Prompting LLMs - Few-shot prompts
J. Results: Misaligned Instruction-Response
K. Results: Ambiguous Impact of Distractors on Reasoning
L. Results: Models Prioritize Contextual Knowledge Over Background Knowledge
M Supplementary Figures and N Supplementary Tables
D Domain-specific pipeline for creating NL instances
E Accessing LLMs
To access these LLMs, we use the Mistral AI (mistral), and the open-source weights of the remaining models, available at the HuggingFace Hub[5] repositories:
\
\ • mistralai/Mistral-7B-v0.1
\ • mistralai/Mistral-7B-Instruct-v0.2
\ • mistralai/Mixtral-8x7B-Instruct-v0.1
\ • google/gemma-7b
\ • google/gemma-7b-it
\ • meta-llama/Meta-Llama-3-8B
\ • meta-llama/Meta-Llama-3-8B-Instruct
\ • BioMistral/BioMistral-7B
\ The pretrained LLM weights are used through the transformers[6] python library. All models were loaded with standard configurations and their respective default tokenizers, using the AutoModelForCausalLM and AutoTokenizer classes. Additionally, the models were loaded with the options devicemap=“auto", torchdtype=“auto" attnimplementation=“flashattention2" and offloadbuffers=True to make the best use of GPU resources available.
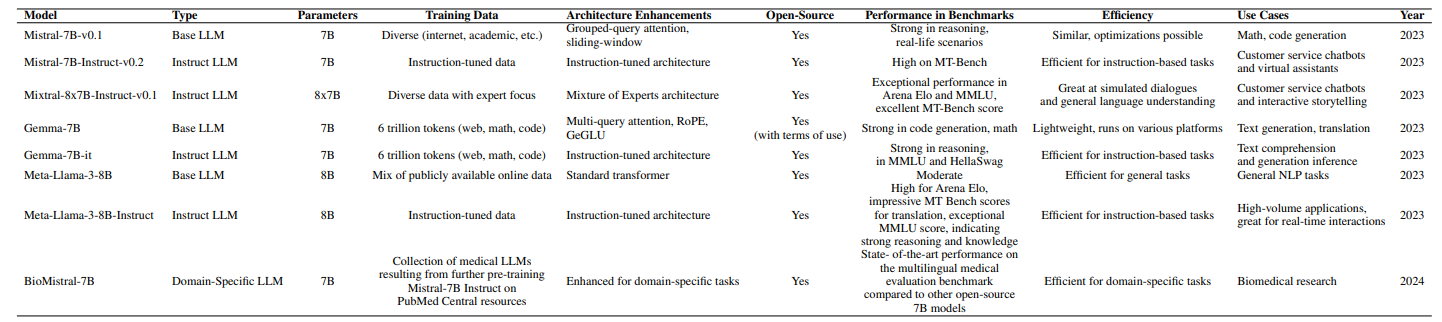
\ For the instruction models, the inputs were passed through each model’s chat template (with tokenizer.applychattemplate), so that they would follow the appropriate prompt format. Responses were cleaned of special symbols for evaluation. Table 3 contains the relevant characteristics of all analyzed models.
\
:::info Authors:
(1) Magdalena Wysocka, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom;
(2) Danilo S. Carvalho, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom and Department of Computer Science, Univ. of Manchester, United Kingdom;
(3) Oskar Wysocki, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom and ited Kingdom 3 I;
(4) Marco Valentino, Idiap Research Institute, Switzerland;
(5) André Freitas, National Biomarker Centre, CRUK-MI, Univ. of Manchester, United Kingdom, Department of Computer Science, Univ. of Manchester, United Kingdom and Idiap Research Institute, Switzerland.
:::
:::info This paper is available on arxiv under CC BY-NC-SA 4.0 license.
:::
[5] https://huggingface.co
\ [6] https://huggingface.co/docs/transformers/
What's Your Reaction?









































































































