Mamba Outperforms HyenaDNA in DNA Sequence Modeling
Mamba scales efficiently for DNA modeling, improving perplexity with larger models and longer context lengths. In DNA sequence classification, Mamba outperforms other models, including HyenaDNA, even in challenging tasks involving species with nearly identical genome
:::info Authors:
(1) Albert Gu, Machine Learning Department, Carnegie Mellon University and with equal contribution;
(2) Tri Dao, Department of Computer Science, Princeton University and with equal contribution.
:::
Table of Links
3 Selective State Space Models and 3.1 Motivation: Selection as a Means of Compression
3.2 Improving SSMs with Selection
3.3 Efficient Implementation of Selective SSMs
3.4 A Simplified SSM Architecture
3.5 Properties of Selection Mechanisms
4 Empirical Evaluation and 4.1 Synthetic Tasks
4.4 Audio Modeling and Generation
4.5 Speed and Memory Benchmarks
\ A Discussion: Selection Mechanism
D Hardware-aware Algorithm For Selective SSMs
E Experimental Details and Additional Results
4.3 DNA Modeling
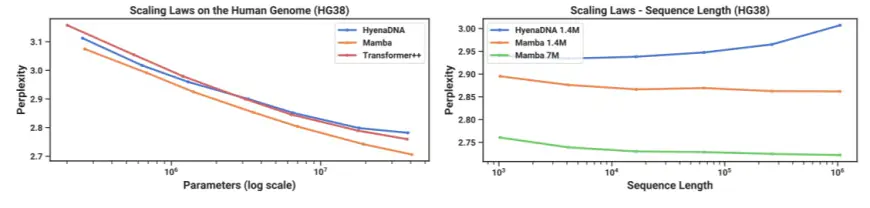
Motivated by the success of large language models, there has been recent exploration into using the foundation model paradigm for genomics. DNA has been likened to language in that it consists of sequences of discrete tokens with a finite vocab. It is also known for requiring long-range dependencies to model (Avsec et al. 2021). We investigate Mamba as a FM backbone for pretraining and fine-tuning in the same setting as recent works on long-sequence models for DNA (Nguyen, Poli, et al. 2023). In particular, we focus on two explorations of scaling laws across model size and sequence length (Figure 5), and a difficult downstream synthetic classification task requiring long context (Figure 6).
\ For pretraining, we largely follow a standard causal language modeling (next token prediction) setup for the training and model details (see also Appendix E.2). For the dataset, we largely follow the setup of HyenaDNA (Nguyen, Poli, et al. 2023), which uses the HG38 dataset for pretraining consisting of a single human genome with about 4.5 billion tokens (DNA base pairs) in the training split.
\ 4.3.1 Scaling: Model Size
\ In this experiment, we investigate the scaling properties of genomics foundation models with various model backbones (Figure 5 Left).
\

\
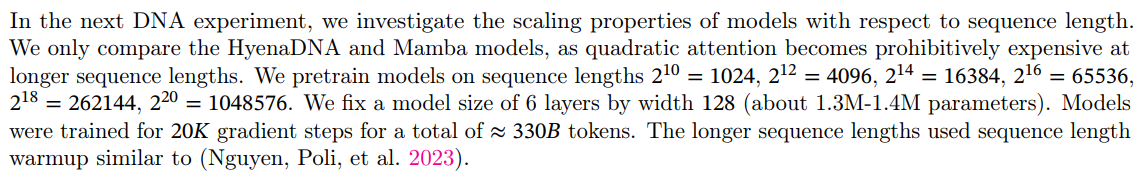
\ Results. Figure 5 (Left) shows that Mamba’s pretraining perplexity improves smoothly with model size, and that Mamba scales better than both HyenaDNA and Transformer++. For example, at the largest model size of ≈ 40M parameters, the curve shows that Mamba can match the Transformer++ and HyenaDNA models with roughly 3× to 4× fewer parameters.
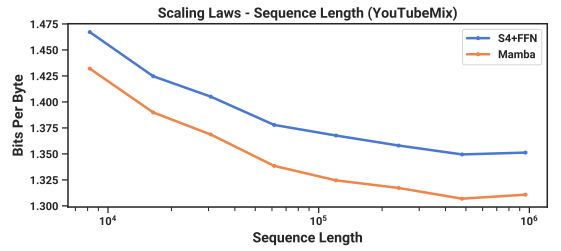
\ 4.3.2 Scaling: Context Length
\
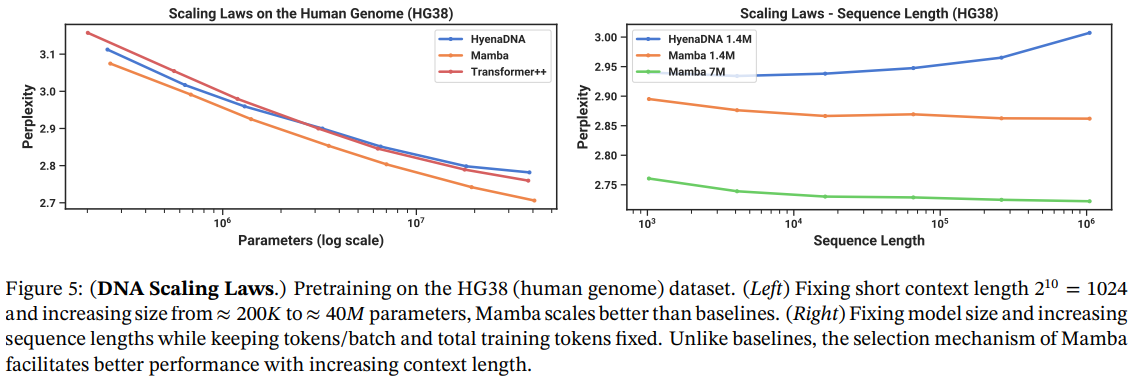
\ Results. Figure 5 (Right) shows that Mamba is able to make use of longer context even up to extremely long sequences of length 1M, and its pretraining perplexity improves as the context increases. On the other hand, the HyenaDNA model gets worse with sequence length. This is intuitive from the discussion in Section 3.5 on properties of the selection mechanism. In particular, LTI models cannot selectively ignore information; from a convolutional perspective, a very long convolution kernel is aggregating all information across a long sequence
\
\
\
\ which may be very noisy. Note that while HyenaDNA claims to improve with longer context, their results do not control for computation time.
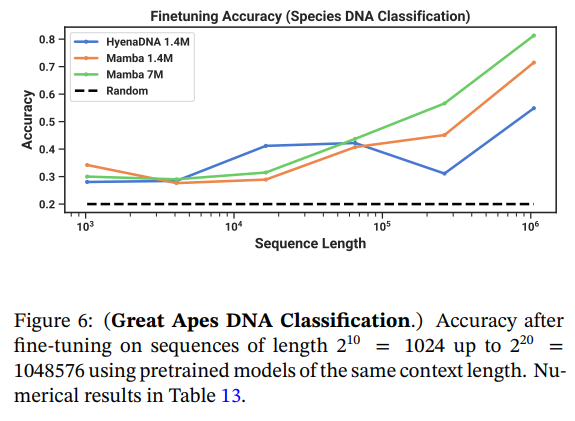
\ 4.3.3 Synthetic Species Classification
\ We evaluate models on a downstream task of classifying between 5 different species by randomly sampling a contiguous segment of their DNA. This task is adapted from HyenaDNA, which used the species {human, lemur, mouse, pig, hippo}. We modify the task to be significantly more challenging by classifying between the five great apes species {human, chimpanzee, gorilla, orangutan, bonobo}, which are known to share 99% of their DNA.
\
:::info This paper is available on arxiv under CC BY 4.0 DEED license.
:::
\
What's Your Reaction?









































































































